

Next: LOAD
Up: Prophet Commands
Previous: GRID
During an IMPLANT step, the statistics from the library are used to
calculate the PearsonIV impurity profile of the implanted ELEMENT. If the
range, standard deviation or higher moments of the desired impurity
distribution are specified, these values overwrite the library values used
to calculate the profile. The user can also specify an exponentially
decaying tail, which begins either at BEGINTAIL or the position where the
concentration is half the peak value, to account for the effects of
channeling.
- ELEMENT=(s)
- DOSE=(r)
- ENERGY=(r)
- [RANGE=(r)]
- [SIGMA=(r)]
- [LAT.SIGMA=(r)]
- [SKEWNESS=(r)]
- [KURTOSIS=(r)]
- [TAIL=(r)]
- [BEGINTAIL=(r)]
- [TILT=(r)]
- [FILE=(s)]
- [DSCALE=(r)]
- [PLUSFACTOR=(r)]
The following list itemizes the valid keywords, their units, and their meaning:
ELEMENT: [string]
impurity element to be implanted. Possible values are found in the
library in library/physics/implant.
DOSE: [real ( 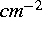 )]
)]
dose of the implant. The dose may be trimmed using the command
dbase create library/physics/silicon/boron/tom.factor rval=0.83
taking boron as an example. This will modify the boron dose to 83% of
the nominal value in every subsequent implant.
ENERGY: [real (keV)]
TILT: [real (degrees)]
tilt of the beam. The model assumes no rotation, only tilt.
RANGE: [real (microns)]
range of the distribution.
SIGMA: [real (microns)]
vertical standard deviation of the distribution.
LAT.SIGMA: [real (microns)]
lateral standard deviation of the distribution.
SKEWNESS: [real]
of the profile (3rd moment).
KURTOSIS: [real]
of the profile (4th moment).
TAIL: [real (microns)]
characteristic length for the exponential tail of the profile.
BEGINTAIL: [real (microns)]
distance from the surface where the tail of the profile should
begin. Default is where the concentration of the implant reaches half
the maximum.
FILE: [string]
name of the file from which the 1D implant statistics is read in.
The file must have two columns, separated by tabs or blank characters.
The first column should give the depth in microns, and the second column
the concentration in 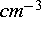 . For a 2D simulation, by default the
lateral sigma is set to equal the vertical sigma value which is
extracted from the 1D implant statistics given by the file. The user
can also set a value for the lateral sigma using the "lat.sigma"
option.
. For a 2D simulation, by default the
lateral sigma is set to equal the vertical sigma value which is
extracted from the 1D implant statistics given by the file. The user
can also set a value for the lateral sigma using the "lat.sigma"
option.
DSCALE: [real]
scaling factor to apply to a profile read from file. Default value is
1.0. Caution: does not apply to profiles specified with dose and
energy, only profiles from file.
PLUSFACTOR: [real]
the implant code to implant a number of interstitials equal
in distribution to the implanted species, and with dose equal to that
of the implanted species multiplied by PLUSFACTOR. The implanted dose
should not exceed the amorphization threshold of silicon; attempts to
do so will be rejected.
Examples:
implant energy=30 dose=1.e16 elem=arsenic
implant element=boron energy=100 dose=1e15 lat.sigma=.021
implant element=arsenic file=my_arsenic lat.sigma=0.1
where file "my_arsenic" is of the form:
0.010 3.0e20
0.012 3.1e20
0.0135 3.3e20
CONCENTRATION
resulting from implant. Either DOSE or
CONCENTRATION may be specified, but not both.
COLUMN.IMSIL
CMODEL
keyword. Should be removed.


Next: LOAD
Up: Prophet Commands
Previous: GRID
Prophet Development
Mon Jul 1 14:45:00 PDT 2002